Research
Our research sits at the interface of the computational and biological sciences. We study gene regulation, usually from an evolutionary perspective. We do so using publicly available experimental data, such as data that describe how regulatory proteins bind DNA to modulate gene expression. We also use computational models of gene regulatory circuits, such as Boolean networks, which allow us to ask general questions about the design constraints, robustness, and evolvability of such circuits.
Adaptive Landscapes
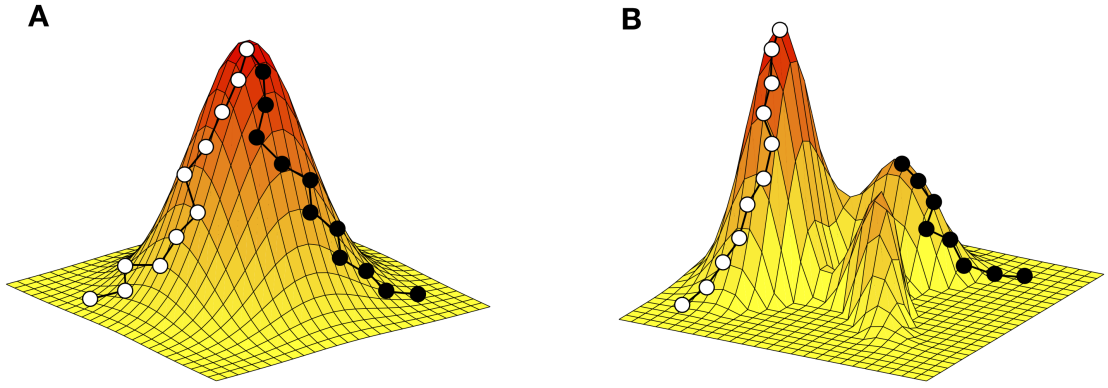
We study empirical adaptive landscapes. This schematic shows how the ruggedness of an adaptive landscape affects the ability of an evolving population to identify the global peak. (A) Single-peaked landscapes pose no obstable to evolution. Both the white and black adaptive trajectories end at the global peak. (B) In a rugged landscape, some adaptive trajectories do not end at the global peak, but rather at a local peak.
Genotype Networks
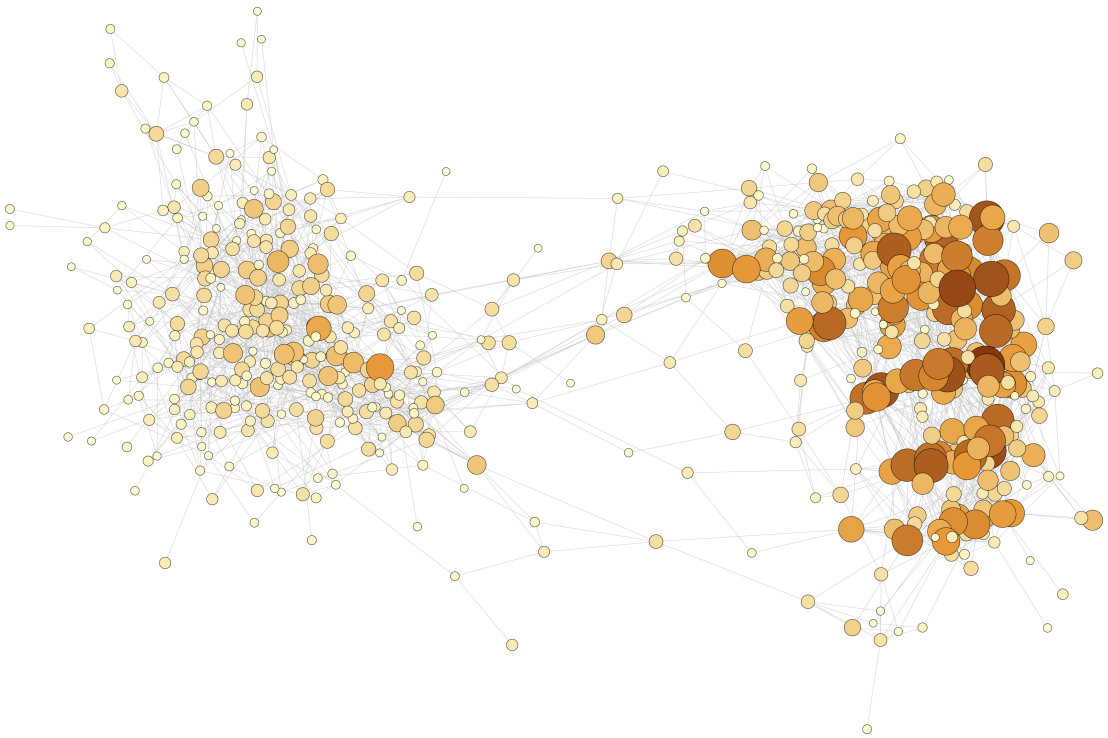
Much of our research centers on the genotype-phenotype map. One common characteristic of a genotype-phenotype map is the existence of genotype networks - sets of mutationally-interconnected genotypes that all have the same phenotype. Shown here is a genotype network of transcription factor binding sites, where each vertex represents a DNA sequence that binds a transcription factor, and edges connect vertices if their corresponding sequences differ by just a single mutation.